Generalized Read-Across (GenRA) Manual
This manual provides an implementation of an algorithmic automated approach to make reproducible read-across predictions of toxicity outcomes from in vivo studies called Generalized Read-across (GenRA) (Shah et al., 2016).
Please use the tabs in the left-hand sidebar or the Next Chapter links at the bottom of the page to navigate through the GenRA manual.
For further information: contact us at genra.support@epa.gov.
Overview
Read-across describes the method of filling a data gap whereby a chemical with existing endpoint information (the source analogue) is used to make a prediction for a ‘similar’ chemical (the target) for the same endpoint. Read-across is a well-established technique used within analogue and category approaches for regulatory purposes (OECD, 2014). ‘Similar’ chemicals are usually identified based on structural similarities, though in practice these analogues are evaluated with respect to other contexts of similarity such as reactivity, physicochemical properties, metabolism and bioactivity.
Read-across is typically performed as an expert driven assessment which poses challenges when scaling to many substances. To address such limitations, Generalized Read-across (GenRA) (Shah et al., 2016) was developed as a data driven approach to facilitate reproducible read-across predictions of toxicity outcomes where performance and uncertainty could be quantified. GenRA relies on a many-to-one read-across approach such that k source analogues are used to make a prediction for a target chemical. .
Within GenRA, source analogues are identified on the basis of structural and/or bioactivity similarity. Future refinements will incorporate other contexts of similarity.
Structural information within GenRA comprises several binary chemical fingerprint representations. Bioactivity information is represented as a binary fingerprint of the ToxCast High Throughput Screening (HTS) assay hitcall outcomes. Herein, we provide some background to the approach and how to navigate through the web-based application (Patlewicz and Shah., 2023).
Background
The original GenRA approach established a baseline in performance for binary predictions of in vivo toxicity. Morgan chemical fingerprints (Rogers and Hahn, 2010), Molecular Access System (MACCS) keys, and topological torsion fingerprints (Nilakantan et al., 1987) were generated for a set of 1778 chemicals taken from ToxCast Phases I-III. Bioactivity fingerprints comprised hitcalls from 821 ToxCast HTS assays. Study-toxicity effect outcomes from over 10 study types, including chronic, sub-chronic, subacute, multigenerational, and developmental toxicity, were taken from ToxRefDB v1.0.
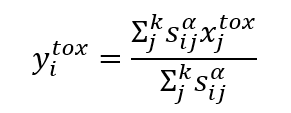
For each pair of chemicals, the chemical similarity, bioactivity similarity, or a hybrid of both chemical and bioactivity similarity were calculated from their vector representations of chemical or bioactivity fingerprints. The predicted read-across toxicity of a chemical was calculated based on the k source analogues using Equation:
Where xjtox is the in vivo toxicity of chemical j. In vivo toxicity outcomes of chemicals were predicted using chemical, bioactivity and hybrid descriptors, that is a = {chm, bio, bc} for k = 2 to 10.
Receiver operating characteristic (ROC) analysis of the predicted and true toxicities was conducted for k-nearest neighbors (where the value of k ranged from 1 to the maximum number of chemicals in the neighborhood), and with a similarity thresholds (where s ranged from the minimum to maximum values of s across all unique pairwise comparisons in the neighborhood). The area under the ROC curve (AUC) was then taken as a measure of performance for a given k and s value. The significance was empirically estimated by constructing a null distribution by permuting the true toxicity 100 times and calculating the fraction of times the AUC was more extreme than what would be observed by chance This was reported as the p-value.
Next chapter: Web Application
